The Zygotic Transition
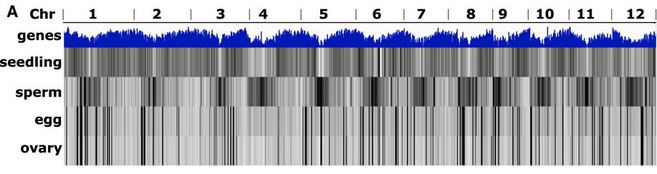 The top row of this whole-genome heat map shows gene density whereas the following rows show the difference between siRNAs from seedling shoot, sperm, egg, and ovary cells.
The top row of this whole-genome heat map shows gene density whereas the following rows show the difference between siRNAs from seedling shoot, sperm, egg, and ovary cells.
Correspondents: Jonathan Gent, PhD and Scott Russel, PhD
Funding: Innovative Genomics Institute & National Science Foundation
The zygote is the ultimate totipotent cell for its powerful ability to develop from a single cell into all different cell types of an organism. Early embryo development is largely dependent on a diverse set of maternal transcripts present in the egg cell before fertilization. The degradation of these maternal transcripts and the initiation of zygotic transcription take place during the Maternal to Zygotic Transition (MZT). A few specific paternal transcripts are delivered by the sperm, some of which serve as triggers for embryo initiation. Egg, sperm and zygote also have very different epigenomes (small RNAs, DNA methylation and histone modifications). We are characterizing the changes in the epigenome starting with the very first cell cycle of the plant embryo, and plan to identify the individual contributions of the maternal and paternal genomes to the zygotic epigenome.
Small RNAs (siRNAs) are a form of transcriptional regulation that cells often use for differentiation. Egg and sperm cells display a wide contrast of siRNA expression compared to vegetative tissues. Further, identification of methylated (silenced) DNA has shown that, in plants, sperm/egg methylated DNA resembles that of vegetative tissue. However, the central cell (which later becomes the endosperm, or nutrients of the seed) experiences widespread demethylation. We hypothesize the contrast in siRNA expression contributes to the differentiation between embryo-bound gametes and the central cell. We aim to uncover this siRNA landscape by sequencing the siRNA transcriptome and DNA methylome of rice plants to thereby understand the parental factors that lead to embryogenesis. Understanding of zygote formation and seed development is crucial for development of synthetic apomixis, the process by which sexual reproduction is replaced by asexual reproduction, bypassing the need for sexual reproduction in the development of seed.
Funding: Innovative Genomics Institute & National Science Foundation
The zygote is the ultimate totipotent cell for its powerful ability to develop from a single cell into all different cell types of an organism. Early embryo development is largely dependent on a diverse set of maternal transcripts present in the egg cell before fertilization. The degradation of these maternal transcripts and the initiation of zygotic transcription take place during the Maternal to Zygotic Transition (MZT). A few specific paternal transcripts are delivered by the sperm, some of which serve as triggers for embryo initiation. Egg, sperm and zygote also have very different epigenomes (small RNAs, DNA methylation and histone modifications). We are characterizing the changes in the epigenome starting with the very first cell cycle of the plant embryo, and plan to identify the individual contributions of the maternal and paternal genomes to the zygotic epigenome.
Small RNAs (siRNAs) are a form of transcriptional regulation that cells often use for differentiation. Egg and sperm cells display a wide contrast of siRNA expression compared to vegetative tissues. Further, identification of methylated (silenced) DNA has shown that, in plants, sperm/egg methylated DNA resembles that of vegetative tissue. However, the central cell (which later becomes the endosperm, or nutrients of the seed) experiences widespread demethylation. We hypothesize the contrast in siRNA expression contributes to the differentiation between embryo-bound gametes and the central cell. We aim to uncover this siRNA landscape by sequencing the siRNA transcriptome and DNA methylome of rice plants to thereby understand the parental factors that lead to embryogenesis. Understanding of zygote formation and seed development is crucial for development of synthetic apomixis, the process by which sexual reproduction is replaced by asexual reproduction, bypassing the need for sexual reproduction in the development of seed.
