Databases
Overview
This is the Sundaresan laboratory database page where you can find data resource updates for our currently funded project ‘Rice Maternal to Zygotic Transition’ (RMZT). Also included are database resources produced during the course of research in the lab since 2001. The links below are organized according to project, with archived materials in the final tab.
Detail
For the RMZT project, we published a paper on characterization of gene expression within the rice gametic cells via RNAseq, and below are links to the final gene differential expression table and a tool to assist in exploring this dataset. Also included here are links to our genome browsers that contain our rice FST genome insertion data as well as data related to small RNA discovery in both rice and maize. The remainder of the menus correspond to specific projects that include:
This is the Sundaresan laboratory database page where you can find data resource updates for our currently funded project ‘Rice Maternal to Zygotic Transition’ (RMZT). Also included are database resources produced during the course of research in the lab since 2001. The links below are organized according to project, with archived materials in the final tab.
Detail
For the RMZT project, we published a paper on characterization of gene expression within the rice gametic cells via RNAseq, and below are links to the final gene differential expression table and a tool to assist in exploring this dataset. Also included here are links to our genome browsers that contain our rice FST genome insertion data as well as data related to small RNA discovery in both rice and maize. The remainder of the menus correspond to specific projects that include:
- Insertional mutagenesis resources
- Arabidopsis miRNA candidate database
- Rice transposon insertion lines
- Small RNA related projects
- Arabidopsis miRNA candidate database
- Cereal small RNA database (rice and maize)
- Various small RNA tools
- Arabidopsis embryo-sac specific genes
-
Rice RNAseq Databases
-
Rice Insertional Mutation Database
-
Plant Small RNA Databases/Tools
-
Arabidopsis Insertional Database (UC Davis Only)
-
Embryo-sac Specific Genes
-
Plant Genome Browser Resources
-
BLAST
-
External Links
<
>
Rice Seed Ordering in the USA:
We ask that seed orders be placed only by a Principal Investigator, or Project Leader.
To receive seeds, two steps are required:
You need to download the APHIS permit, add your specific information , and submit it to APHIS by following the instructions on page 3 of the form.
You will also need an interstate movement notification document (prefilled) that has all of the pertinent information about the rice insertion lines. It covers all of our insertion lines.
Once APHIS receives your notification, they will notify your State agency, and send you an approval. The process should take ten working days to two weeks to complete.
After you receive approval from APHIS, mail or FAX a copy of it to:
Aaditi Gaikwad
Plant Biology
University of California
One Shields Avenue
Davis CA 95616 USA
FAX 530-752-5410
You can then order online, or send in a written order along with the approval document.
We ask that seed orders be placed only by a Principal Investigator, or Project Leader.
To receive seeds, two steps are required:
- Due to the fact that you are ordering transgenic crop seeds, the USDA /APHIS requires you to file an interstate movement notification. We cannot send seed to you in the United States without your having a movement notification document approved by APHIS and submitted to us. (Guidance for this step follows below ).
- Recipient will have to certify that the Pathogen Status Disclaimer has been read and that Recipient will abide by conditions of the Distribution Agreement. (This is done by submission of a seed order)
You need to download the APHIS permit, add your specific information , and submit it to APHIS by following the instructions on page 3 of the form.
You will also need an interstate movement notification document (prefilled) that has all of the pertinent information about the rice insertion lines. It covers all of our insertion lines.
Once APHIS receives your notification, they will notify your State agency, and send you an approval. The process should take ten working days to two weeks to complete.
After you receive approval from APHIS, mail or FAX a copy of it to:
Aaditi Gaikwad
Plant Biology
University of California
One Shields Avenue
Davis CA 95616 USA
FAX 530-752-5410
You can then order online, or send in a written order along with the approval document.
Rice Seed Ordering Internationally:
We are not currently set up to handle online orders from outside the United States.
If you are interested in ordering from outside the U.S., please send an email to Aaditi Gaikwad, [email protected].
We are not currently set up to handle online orders from outside the United States.
If you are interested in ordering from outside the U.S., please send an email to Aaditi Gaikwad, [email protected].
-
Arabidopsis miRNA Candidates
-
Cereal Small RNA Database (CSRDB)
-
Small RNA Target Site Tools
<
>
What is this page?
This page represents a resource of predicted miRNA and precursor candidates for the Arabidopsis genome predicted by the algorithm 'findMiRNA' and is also the supplementary material for the paper titled 'Computational Prediction of miRNAs in Arabidopsis thaliana'. This page is provided to give the scientific community a clean interface to all of our raw computational results. The findMiRNA algorithm has the sensitivity to detect most previously reported miRNAs (see 'The miRNA Registry' at Rfam). It should be noted that other sequences with the potential to form hairpins are also identified by this algorithm such as tRNAs, foldback elements and retrotransposons. Whether a sequence represents a miRNA precursor or not should be determined by users of this site and the data is provided unfiltered.
This page represents a resource of predicted miRNA and precursor candidates for the Arabidopsis genome predicted by the algorithm 'findMiRNA' and is also the supplementary material for the paper titled 'Computational Prediction of miRNAs in Arabidopsis thaliana'. This page is provided to give the scientific community a clean interface to all of our raw computational results. The findMiRNA algorithm has the sensitivity to detect most previously reported miRNAs (see 'The miRNA Registry' at Rfam). It should be noted that other sequences with the potential to form hairpins are also identified by this algorithm such as tRNAs, foldback elements and retrotransposons. Whether a sequence represents a miRNA precursor or not should be determined by users of this site and the data is provided unfiltered.
Important Introductory Notes
Before using this database:
Before using this database to look for potential miRNA target sites in your gene of interest, it is advisable that you already have some evidence suggesting that your gene is regulated by a miRNA. Such evidence might originate from genetic and/or bioinformatic sources. A genetic example would be a silent base substitution in the coding region (i.e. does not result in an amino acid substitution) that behaves as a dominant allele. This point mutation may be present in a miRNA target site and thus blocking negative regulation of the gene by a miRNA. A bioinformatic example would be the presence of a conserved ~20 nt sequence across a number of partially diverged genes and this may represent a conserved miRNA target site.
How to search the database:
To use this database, go to the search form and enter the transcript ID (as defined by TAIR) for your gene of interest. The default filter settings will reduce the data by 96 percent and results in an average of 3 hits per transcript. The search page input allows for the results to be limited to those precursors that have a presumptive homologue in rice as found by precExtract, an internally developed program that searches for miRNA homologues in other genomes. The sequence results can be shown as either as RNA or DNA, with RNA as the default option.
miRNA precursor families:
Many of the easily identifiable miRNA precursor families are likely to have already been discovered during the course of our work. However, it is still possible that other precursor families do exist, and therefore the input page has been set up to allow users to limit the output to records for which the candidate miRNAs overlap single target sites on the transcript, a property of precursor families. These overlapping candidate precursor sequences can also be distinguished from the other data without selecting the filter, as the target site nucleotide range is shown in green text for these candidates. After the results are returned, individual candidate precursor sequences can then be selected by checking the boxes on the left and aligned using the provided Clustalw alignment option at the base of the page (more computationally intense alignments were performed for the publication using T_COFFEE). This enables the detection of the characteristic divergence pattern that is often observed for precursor families, where the miRNA and miRNA* sequences are more conserved than the intervening sequence of the hairpin.
Target site conservation:
In addition to looking for the characteristic divergence pattern seen in precursor families, searching for conservation of target sites within related transcripts within Arabidopsis or between Arabidopsis and another species is highly recommended. Clear evidence of the presence of a conserved target site is the strongest indicator of a bona fide miRNA. Another strong evidence point is the presence of more than one copy of the same target site within a single transcript. This is consistent with cooperative regulation of the transcript by the presumptive miRNA. We recommend using PATMATCH/WU-BLAST/BLASTN at TAIR to help identify potentially conserved target sites in plant transcripts allowing a few mismatches to account for G-U pairing that may occur between the miRNA and its target. It is suggested that precursor sequences are aligned to plant genomic DNA using BLAST at NCBI as this will often identify noise such as tRNAs and can be used to detect the presence of potential miRNA precursors in other plant genomes. Subsequent hairpin prediction for these potential precursor sequences can then be performed using either RNAFold or Mfold.
Bioinformatic Credits
Alex Adai, Cameron Johnson, Varun Manocha
If you use our data and/or software in your research, please cite this web resource, and our paper:
Computational Prediction of miRNAs in Arabidopsis thaliana.
Alex Adai, Cameron Johnson, Sizolwenkosi Mlotshwa, Sarah Archer-Evans, Varun Manocha, Vicki Vance, and Venkatesan Sundaresan. Genome Research 15:78-91, 2005
Addendum - Identification of miR395 targets by findMiRNA.
Acknowledgements
We are extremely grateful to Edward Marcotte and the Marcotte Lab for previously hosting and supporting this site. We would also like to thank Andrej Sali of the University of California San Francisco for providing access to critical computational resources for the most recent runs of findMiRNA.
Small RNAs link to predicted targets
Bioinformatics Summary - Current Status:
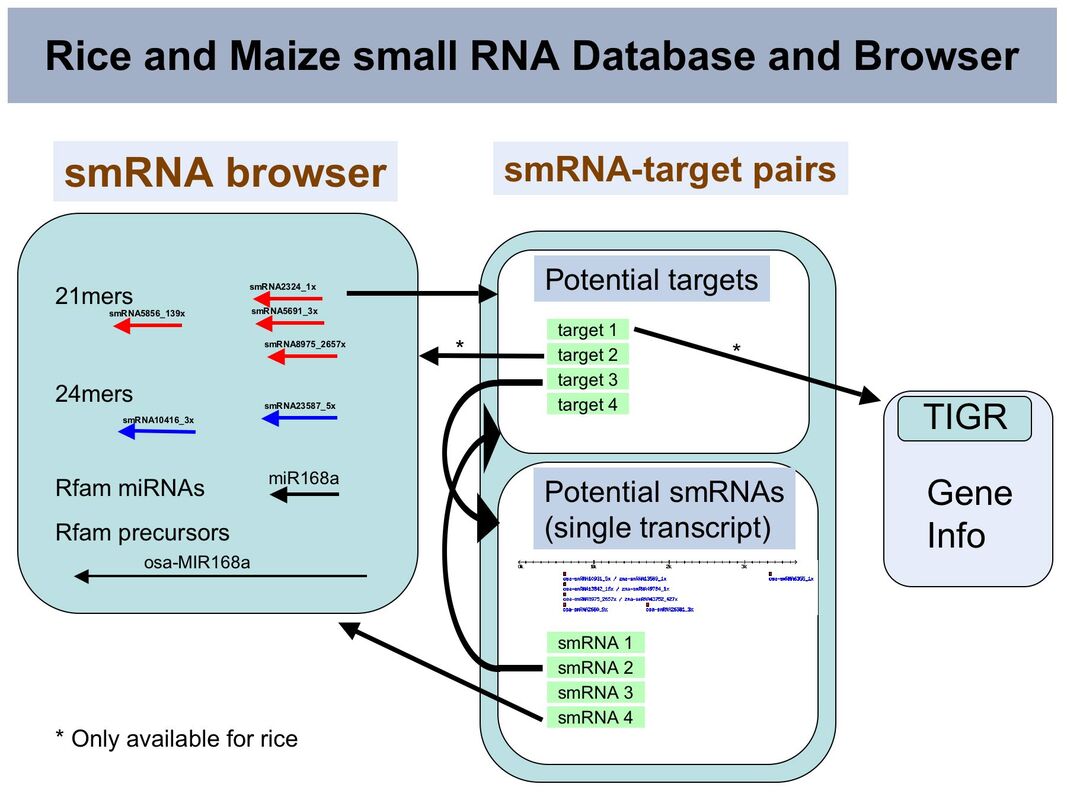
Plant small RNAs, which include miRNAs, siRNAs and trans-acting siRNAs (ta-siRNAs), represent a class of regulatory molecule that is increasingly being seen as a significant component of epigenetic processes as well as an important component of gene networks involved in development and in homeostasis. Here we present a bioinformatic resource for cereal crops consisting of large-scale datasets of maize and rice small RNA sequences generated by 454 Life Science sequencing. The small RNA sequences have been mapped to the rice genome and available maize genome sequence and are presented in two genome browser datasets using the Generic Genome Browser (Lincoln Stein). Potential target sequences representing mature mRNA sequences have been predicted using the FASTH software from the Zuker lab. and access to the resulting small RNA target pair (SRTP) dataset has been made available through a mysql based relational database. Within the genome browser the small RNAs have links to the SRTP database that will return a list of potential targets. The SRTP database may also be searched independently using both small RNA and target transcript queries. Data linking and integration is the main focus of this interface and to this aim links are present in the SRTP results pages back to the browser and the SRTP database as well as external sites. The resource will be updated as more sequences become available.
Plant small RNAs, which include miRNAs, siRNAs and trans-acting siRNAs (ta-siRNAs), represent a class of regulatory molecule that is increasingly being seen as a significant component of epigenetic processes as well as an important component of gene networks involved in development and in homeostasis. Here we present a bioinformatic resource for cereal crops consisting of large-scale datasets of maize and rice small RNA sequences generated by 454 Life Science sequencing. The small RNA sequences have been mapped to the rice genome and available maize genome sequence and are presented in two genome browser datasets using the Generic Genome Browser (Lincoln Stein). Potential target sequences representing mature mRNA sequences have been predicted using the FASTH software from the Zuker lab. and access to the resulting small RNA target pair (SRTP) dataset has been made available through a mysql based relational database. Within the genome browser the small RNAs have links to the SRTP database that will return a list of potential targets. The SRTP database may also be searched independently using both small RNA and target transcript queries. Data linking and integration is the main focus of this interface and to this aim links are present in the SRTP results pages back to the browser and the SRTP database as well as external sites. The resource will be updated as more sequences become available.
N.B. In keeping with the predominant notation in the small RNA community, the column formally labeled as 'unique' in the above table is now labelled as 'distinct', and refers to the sequence and not to a map position. That is, the total nucleotide sequences can be reduced to a set of distinct sequences, some of which may be mapped to multiple places within the genome sequence.
Tab under construction. Check again later!
Identification of female gametophyte-specific genes using oligonucleotides microarrays
Since gametophyte development occurs within the sporophyte, the isolation of RNA from developing gametophytes without contamination with the maternal tissues represents a technical challenge. We previously described a gene called SPOROCYTELESS (SPL) required for initiation of gametophyte development in Arabidopsis (Yang et al. 1999). Plants that are homozygous for the spl mutation have ovules that appear normal except that they do not contain embryo sacs, and the nucellus does not degenerate. Therefore, the spl mutant provides an opportunity to identify female gametophyte-specific genes by expression profiling. Oligonucleotide microarrays are being used to identify genes specific for the female gametophyte by comparison of the expression profiles of wild-type ovules (embryo-sac + ) with spl mutant ovules (embryo sac - ).
Click on title below to link to reprint from Plant Physiology
Gametophyte Transcriptome of Arabidopsis by Comparative Expression Profiling
Plant Physiology 2005 : 105067314
Gametophyte Transcriptome of Arabidopsis by Comparative Expression Profiling
Plant Physiology 2005 : 105067314
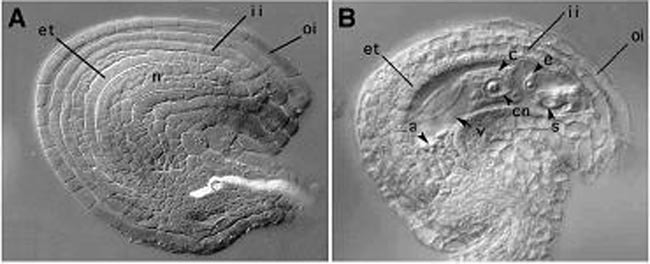
A, Image of spl ovule at maturity showing the complete absence of the embryo sac but presence of all maternal cell types. B. WT ovule with embryo sac for comparison.
-
Arabidopsis
-
Rice
-
Maize
-
Sequence Search
<
>
Tab under construction. Check again later!
Tab under construction. Check again later!