Genomics of Host-Microbiome Interactions in Rice
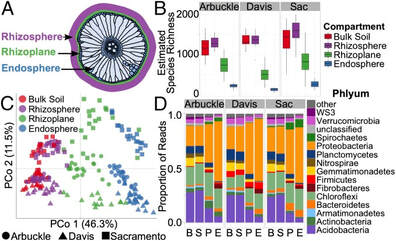 The plant root is typically divided into three separate compartments: the rhizosphere, rhizoplane, and endosphere. These not only display differences in their microbiome between each other, but their differences between soil environments are also unique
The plant root is typically divided into three separate compartments: the rhizosphere, rhizoplane, and endosphere. These not only display differences in their microbiome between each other, but their differences between soil environments are also unique
Correspondents: Jonathan Eisen, PhD; David Mackill, PhD; Merle Anders, PhD
Funding: National Science Foundation
Plants, much like humans and other animals, harbor rich communities of bacteria, including some with potentially beneficial traits. Soil microbes in close proximity to plant roots (the rhizosphere) have been shown to have direct effects on plant growth. Using rice (Oryza sativa) as a model, we are answering questions based around how plants recruit and moderate their associated root microbiomes. Unraveling the composition and maintenance of the rice microbiome is not only important for bolstering our basic understanding of host-microbe interactions: it is of great agronomic and ecological importance as well.
One aspect of rice-microbe interactions we study is the potential protective value of the microbiome when the plant undergoes drought stress. Understanding and applying this value is crucial for not only maintaining but increasing future agricultural yields to feed a growing human population during times of increased drought. These broad surveys of microbes that associate with plants during drought stress allow us to identify potential strains that could mitigate that stress. We have also isolated many microbes that show changes under drought and are exploring if they have the potential to benefit rice plants under drought stress.
Funding: National Science Foundation
Plants, much like humans and other animals, harbor rich communities of bacteria, including some with potentially beneficial traits. Soil microbes in close proximity to plant roots (the rhizosphere) have been shown to have direct effects on plant growth. Using rice (Oryza sativa) as a model, we are answering questions based around how plants recruit and moderate their associated root microbiomes. Unraveling the composition and maintenance of the rice microbiome is not only important for bolstering our basic understanding of host-microbe interactions: it is of great agronomic and ecological importance as well.
One aspect of rice-microbe interactions we study is the potential protective value of the microbiome when the plant undergoes drought stress. Understanding and applying this value is crucial for not only maintaining but increasing future agricultural yields to feed a growing human population during times of increased drought. These broad surveys of microbes that associate with plants during drought stress allow us to identify potential strains that could mitigate that stress. We have also isolated many microbes that show changes under drought and are exploring if they have the potential to benefit rice plants under drought stress.
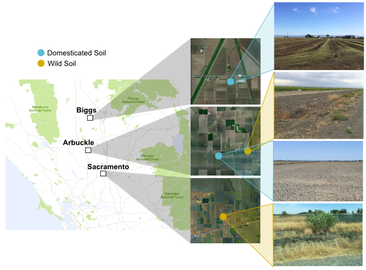 Location variances based on how long ago fields were used for cultivation exemplifies how "domestication" of soil can affect the microbiome
Location variances based on how long ago fields were used for cultivation exemplifies how "domestication" of soil can affect the microbiome
Another focus of our research deals with understanding how the microbiome changes over time. Previous work attempting to increase yield by application of microbes has failed as plants, like humans, experience a change in their microbiome composition as they age. Understanding these seasonal changes in microbiome could help us understand why previous work has yet to yield significant improvements and, in the future, inform how best to apply microbes to crop plants given their age-dependent microbiome.
Further, our research also concerns the production of methane, a greenhouse gas 25 times as potent as carbon dioxide, in the cultivation of rice as rice paddies account for 15 – 20% of global methane emissions. Methane is produced by methanogenic archaea that feed on organic material in the paddy soil. The produced methane is taken up by the rice plant and emitted through a specialized gas vascular system. Interestingly, the rice plant provides an environment conducive for methane-utilizing bacteria by partially oxygenating the soil adjacent to the roots, providing a substrate for methane oxidation. As this understanding is crucial for mitigating methane emissions resulting from rice cultivation, we seek to elucidate mechanisms for how a rice plant moderates levels of these methane-utilizing bacteria.
We also study how continually growing rice season after season affects the microbiome. Distinctions between microbiota occur between soil which has previously grown crop plants and "wild" soil. This distinction contributes to the "domestication" of soil that results in decreased yield from plants grown, likely as a trade-off between pathogenic infection of plants and yield. We aim to elucidate this role by characterization of microbiota existing in the soil.
Finally, we aim to not only profile the microbes that associate with rice plants, but understand the functions and interactions these communities and their host have at a molecular level as well. We have developed sterile growth conditions for rice plants to profile the transcriptional regulation that occurs when plants are interacting with a microbial community. As well, this system allows for rice plants to be inoculated with a consortia of microbes to not only profile how individual microbes interact with a rice plant, but how those individual microbial interactions are affected by the addition of other microbes. This method represents a major step forward in understanding these interactions at the genetic level and provides vital background for understanding of non-pathogenic plant-microbial interactions.
Further, our research also concerns the production of methane, a greenhouse gas 25 times as potent as carbon dioxide, in the cultivation of rice as rice paddies account for 15 – 20% of global methane emissions. Methane is produced by methanogenic archaea that feed on organic material in the paddy soil. The produced methane is taken up by the rice plant and emitted through a specialized gas vascular system. Interestingly, the rice plant provides an environment conducive for methane-utilizing bacteria by partially oxygenating the soil adjacent to the roots, providing a substrate for methane oxidation. As this understanding is crucial for mitigating methane emissions resulting from rice cultivation, we seek to elucidate mechanisms for how a rice plant moderates levels of these methane-utilizing bacteria.
We also study how continually growing rice season after season affects the microbiome. Distinctions between microbiota occur between soil which has previously grown crop plants and "wild" soil. This distinction contributes to the "domestication" of soil that results in decreased yield from plants grown, likely as a trade-off between pathogenic infection of plants and yield. We aim to elucidate this role by characterization of microbiota existing in the soil.
Finally, we aim to not only profile the microbes that associate with rice plants, but understand the functions and interactions these communities and their host have at a molecular level as well. We have developed sterile growth conditions for rice plants to profile the transcriptional regulation that occurs when plants are interacting with a microbial community. As well, this system allows for rice plants to be inoculated with a consortia of microbes to not only profile how individual microbes interact with a rice plant, but how those individual microbial interactions are affected by the addition of other microbes. This method represents a major step forward in understanding these interactions at the genetic level and provides vital background for understanding of non-pathogenic plant-microbial interactions.



